Research Projects and goals
The overall aims of research in my laboratory are to better understand, at the molecular and biochemical level, the following processes:
*Development of novel antifungals and elucidation of their mode of action
*Identification of novel virulence genes in A. fumigatus.
Major Research Findings (2001-date)
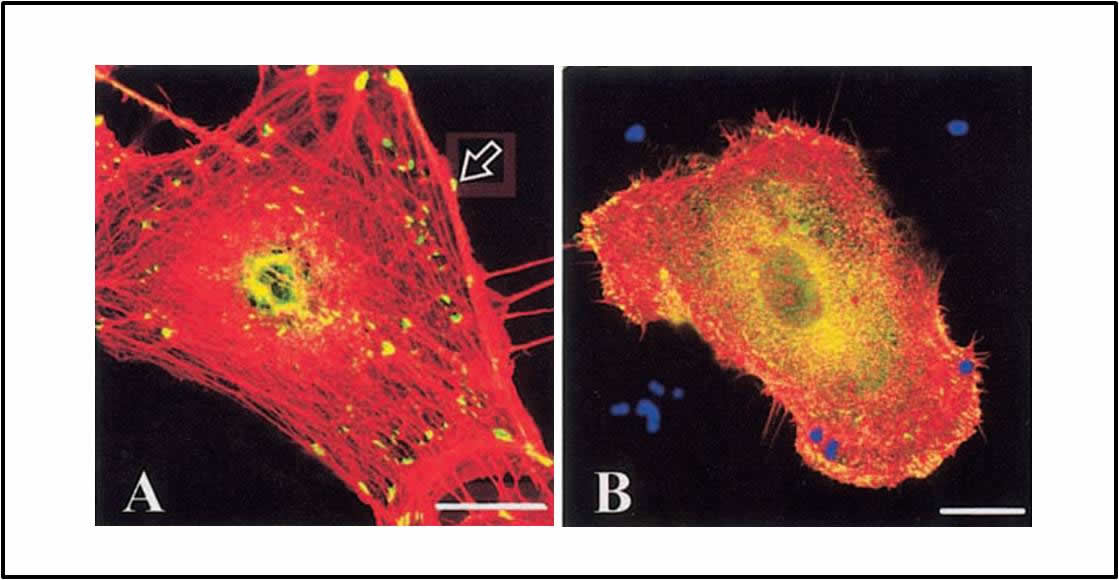
1. Aspergillus fumigatus pathogenesis and the role of secreted proteases. We have shown for the first time that A. fumigatus secreted proteases are involved in the disruption of the actin fiber cytoskeleton and loss of focal adhesion sites in infected lung cells (Kogan et al. 2004). These results suggest that combining protease inhibitors with current antifungals may reduce damage at the site of infection.
Recently we have analyzed changes in phosphorylation of signaling molecules in A. fumigatus infected lung cells. We have shown activation of the MAPK and PKC pathways. Transcriptomic analysis using microarrays revealed transcriptional activation of genes in the NF-kapp B pathway (Sharon et al. 2011). We have generated a protease null A. fumigatus mutant in which we deleted the transcriptional regulator PrtT (Sharon et al. 2009; Hagag et al. 2012). However, this mutant retained full virulence in a mouse model of Aspergillus infection.
2. Characterization of A. fumigatus cell-wall proteins. We have used a bioinformatics approach to identify putative A. fumigatus cell-wall proteins. Deletion of one of them , ECM33, resulted in a phenotype characterized by premature germination and hypervirulence (Romano et al. 2006). We have also used bioinformatics to identify all tandem-repeat rich genes in A. fumigatus. Deletion of one of them, Afu3g08990, encoding a cell-wall protein resulted in premature germination and reduced adherence (Levdansky et al. 2007) weakened cell wall and reduced resistance to phagocytosis (Levdansky et al. 2010). Analysis of the CFEM-motif containing wall proteins showed deletion of all three genes resulted in reduced resistance to cell wall stressors but no change in virulence (Vaknin et al. 2014).

3. Aspergillus fumigatus hypoxia adaptation: A. fumigatus forms a hypoxic microenvironment during lung infection and survives low oxygen availability by activating the srbA signaling pathway. We have used genetic screening to identify a novel protein RbdA involved in this pathway. RbdA is a transmembranal protease that activates the transcription factor srbA by cleavage, freeing it to enter the nucleus (Vaknin et al. 2016). Deletion of rbdA leads to inability of the fungus to grow under hypoxia and complete loss of virulence in infected mice.
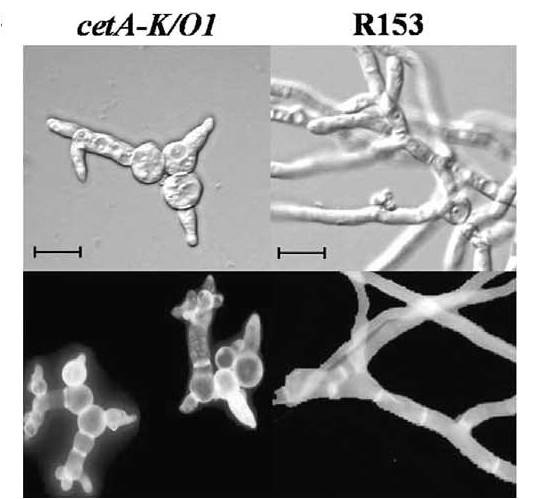
4. Aspergillus nidulans germination: We have used subtractive hybridization to identify transcripts preferentially accumulated in dormant conidia (Osherov et al. 2001). The role of one of these, a thaumatin-like gene called cetA, and its closely related homolog calA, have been studied in detail (Greenstein et al. 2005, Belaish et al. 2007). Both genes encode for small (23 kd) secreted proteins which can be visualized in the cell wall, extracellular matrix and supernatant. While deletion of each gene alone does not impair fungal growth, deletion of both genes results in the generation of fragile conidia which lyse during germination. Our group is the first to characterize the function of these genes in fungi. Work in A. fumigatus has shown that calA is an invasin
5. Discovery of Novel antifungals: We have prepared and characterized a conditional PKC strain of A. nidulans (Ronen et al. 2007). This mutant is highly sensitive to cell wall damaging agents under PKC-supressive conditions. We utilized it to screen a large compound library to identify those with cell-wall damaging activity (Mircus et al. 2009). We further identified the in vitro and in vivo activity of 2 interesting antifungal compounds, CANBEFs that inhibit fungal protein synthesis (Mircus et al. 2015), hemofungin that inhibits fungal heme synthesis (Ben-Yaakov et al 2016) and bromoguinol that activates fungal programmed cell death (Ben-Yaakov et al. 2017).
In collaboration with the Mirelman group (Weizmann), we developed highly effective anti-aspergillus monoclonal antibodies conjugated to the enzyme allinase. This enzyme, cloned from garlic, produces the toxic reactant allicin that kills the fungus. The conjugate showed high efficacy in vitro and in vivo (Appel et al. 2010).
In collaboration with the Shai group (Weizmann), we developed highly effective anti-fungal trivalent ultrashort lipopetides . They showed high efficacy in vitro and in vivo (Arnusch et al. 2012 a,b).
In collaboration with the Benhar group (TAU)we developed a novel non-toxic and highly soluble derivative of Amphotericin B, a known antifungal drug. It was effective and non toxic in vivo in mice infected with Aspergillus fumigatus (Halperin et al. 2016).
6. Discovery of novel antifungal drug targets: In collaboration whith Prof. Brakahage and the Jena group we have identified a novel rhomboid protease RbdA, involved in survival under lung hypoxic conditions (Vaknin et al. 2016). In collaboration whith Prof. Nancy Keller (Wisconsin) we have identified the copper resonse pathway in A. fumigatus, composed of a sensor transcription factor AceA and a downstream Cu exporter CrpA. Loss of these genes results in reduced virulence of the fungus during infection (Wiemann , Perevitsky et al. 2017). In collaboration with Prof. Hubertus Haas (Innsbruk), we have identified key biosynthetic pathways involved in A. fumigatus virulence, including the biosynthesis of pantothenic acid, riboflavin and leucine (Dietl, Meir et al. 2018; Orasch et al 2019)
7. Novel drug combinations against Aspergillus species: *We have shown that the combination of caspofungin, which inhibits fungal cell wall synthesis, and itraconazole, which inhibits fungal ergosterol biosynthesis is synergistic in vitro (Shalit et al., 2003)
*We have used, for the first time, a chemogenomic approach to identify novel antifungal drug combinations (Markovitz et al., 2004). We analyzed, the S. cerevisiae genome deletion library to identify new genes affecting susceptibility to the novel antifungal drug caspofungin. Our results suggested that the combination of caspofungin with inhibitors of the PKC-dependent cell wall repair pathway may result in synergy. We therefore tested the combination of caspofungin and the PKC inhibitor staurosporine and found them to be strongly synergistic in both S. cerevisiae and several pathogenic Aspergillus species, including A. fumigatus. These results suggest a novel approach to antifungal combination therapy, through the development of fungal-specific PKC inhibitors used in combination with drugs that inhibit fungal cell wall biosynthesis such as caspofungin.
* The antibacterial drug sulfamethoxazole (SMX) inhibits fungal growth through inhibition of folate biosynthesis. Moreover, A. fumigatus mutants defective in this pathway are avirulent. We therefore tested the combination of SMX with several antifungal drugs. Using checkerboard, time kill and microscopic analysis, we discovered that the combination of SMX and caspofungin is synergistic in vitro (Yekutiel et al. 2004).
8. Novel Methodologies developed in the lab: We have developed a novel, rapid transposon-based method for disrupting genes in A. nidulans and A. fumigatus (Jadoun et al., 2004). We have used this method successfully to obtain, for the first time, A. fumigatus argB auxotroph strains that can be of use for the genetic analysis of this fungus. We have also used this method to disrupt the AfuEcm33 and cetA genes described above.

