|
Microbial Thiol-Disulfide Redox Metabolism
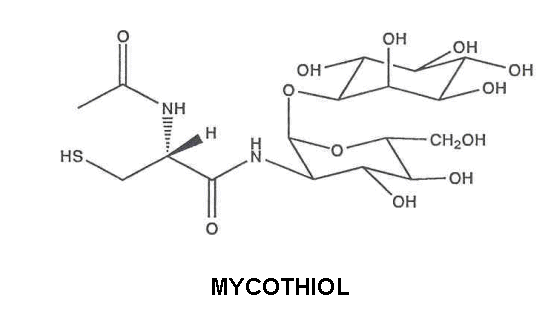
Enzymatic thiol-disulfide exchange reactions play a key role in many metabolic processes. Our interest focuses on the nature and activity of such redox systems in the biology of gram-positive bacteria that lack glutathione, in particular in their growth and development, for the production of secondary metabolites and as targets for creating new antibacterial drugs. We have shown that Streptomyces and Mycobacterium and many other members of the high G+C branch of the actinomycetes contain an unusual low molecular weight thiol, termed mycothiol (MSH), that is present in millimolar intracellular concentrations. MSH may play an analogous role to that of glutathione - once thought to be ubiquitous in living organisms - in maintaining the reducing thiol-disulfide status of the cytoplasm, as a hydrogen donor for essential cellular processes, and in the elimination of reactive
oxygen species (33,38, 41). In Streptomyces and in some other gram-positive bacteria, including Staphylococcus aureus the major thiol-disulfide reductase is thioredoxin/thioredoxin reductase (32,34). We are currently examining the regulation of the thiol redox systems in these bacteria in response to disulfide stress (51) and to other environmental stress agents as well as their potential as targets for the development of new classes of antibacterial drugs (47).
Genetic
and Biochemical Studies of Ribonucleotide Reductase
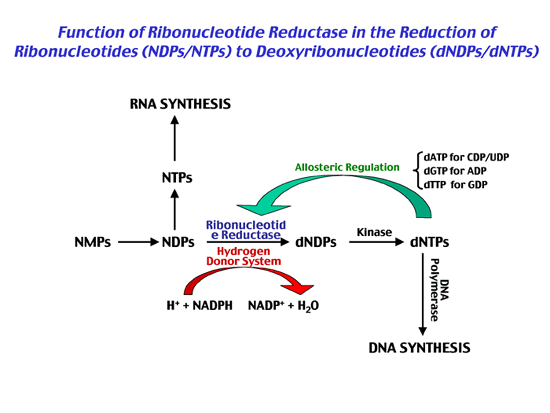
Ribonucleotide reductases (RNR) are essential enzymes that catalyze the reduction of ribonucleotides to deoxyribonucleotides for DNA replication and repair. We have cloned and sequenced the Streptomyces nrdAB and nrdJ genes encoding class Ia oxygen-dependent and class II oxygen-independent ribonucleotide reductases and shown that they are differentially transcribed during vegetative growth (49). Studies are directed towards determining the cellular roles of the two RNRs. Similar studies with Staphylococcus aureus reveal that they contain genes coding for class Ib aerobic and class III anaerobic ribonucleotide reductases that are regulated in response to changes in oxygen tension (50). We are currently investigating the molecular mechanisms that control the expression of these genes in anaerobiosis.
Structure-Function
Studies of Isopenicillin N Synthase
Isopenicillin N synthase is a non-heme ferrous iron dependent dioxygenase that plays a central role in penicillin biosynthesis, catalyzing a unique reaction in which a linear tripeptide, delta-(L-alpha-aminoadipoyl)-L-cysteinyl-D-valine (ACV), is oxidized to form a bicyclic beta-lactam antibiotic (56). Our studies are directed towards elucidating the nature of the substrate binding and catalytic active-site domains, thereby enabling a better understanding of the reaction mechanism. Site-directed mutagenesis and sequence analysis were used to identify the endogenous active-site ligands and led to the assignment of a novel motif for iron binding (40, 43, 46). The crystal structure of the Streptomyces jumonjinensis IPNS and of a substrate-inhibitor complex were recently determined and afford new insights into the mechanism of the cyclization reaction.
 Recent Research Support from Academic Institutions 1994-97 US-Binational Science Foundation Structure Function of Isopenicillin N Synthase 1995- 98 German-Israel Binational Science Foundation Management of Oxidative Stress in the Listerial Host Cell Interaction 1997- 00, 2000- 03 The Israel Science Foundation Mechanism of Action of Isopenicillin N Synthase 2001-2004 The Israel Science Foundation Regulation of the Staphylococcus aureus ribonucleotide reductase genes at the aerobic-anaerobic interface top Cohen home microbiology home |